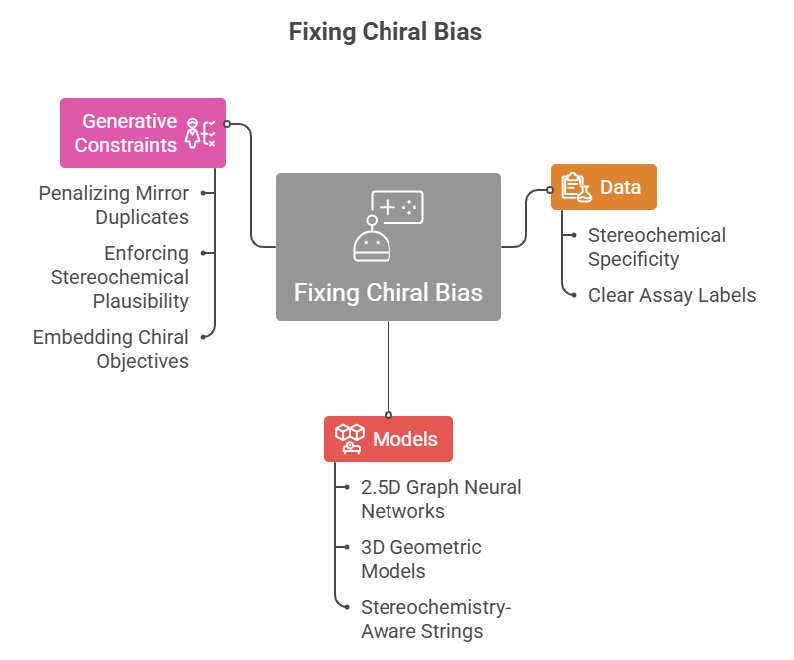
❓ How do we move from recognizing chiral bias to responsibly fixing it?
On Visuals and Stereochemical Truth
Some illustrations in this series are generated or assisted by AI to support conceptual understanding. These visuals are intentionally simplified and should not be read as stereochemically rigorous or chemically exact representations. Wherever stereochemical fidelity matters, it is addressed explicitly in the discussion—because in chemistry, especially in chiral systems, intuition must always yield to structure.
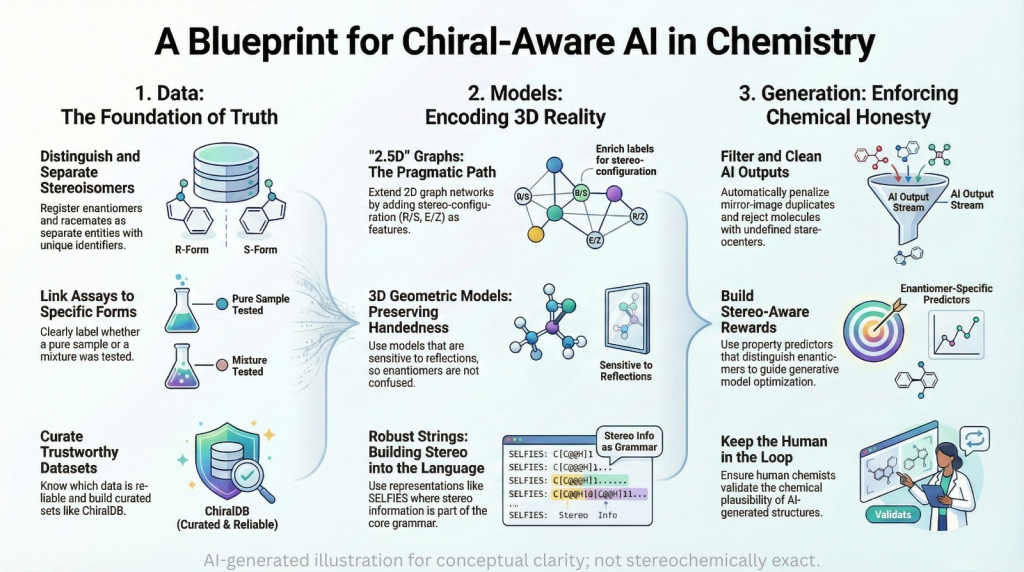
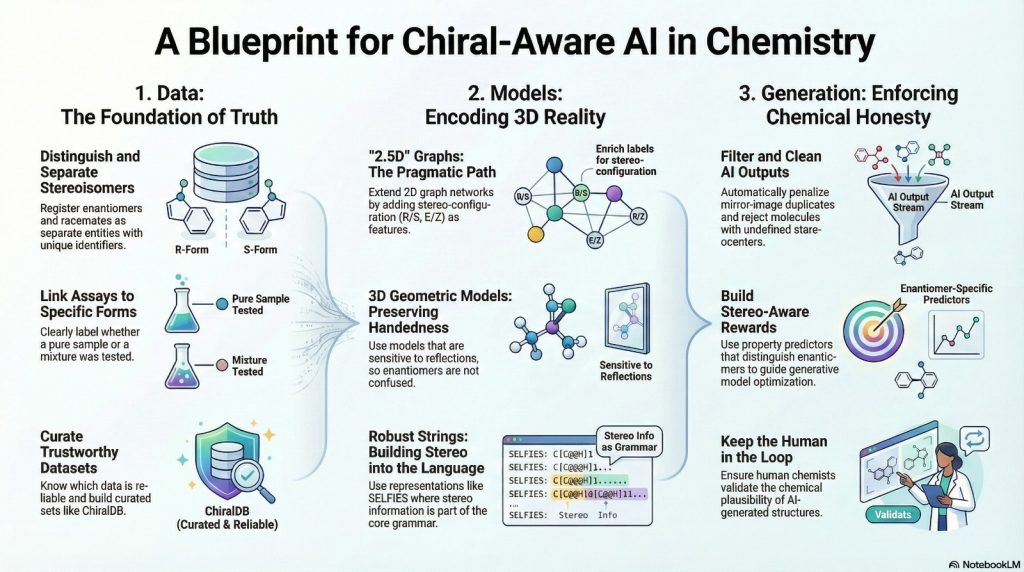
The earlier parts of this series framed chiral bias as a mismatch between the three dimensional nature of chemistry and the flattened representations used in many AI pipelines. This episode is about remedies. It groups the solution into three layers: data, representations and architectures, and generative constraints.
On the data side, the goal is simple to state and hard to execute: every record should refer to a specific stereochemical form, and assay labels should be clear about what was actually tested. On the model side, we look at two and a half dimensional graph neural networks, three dimensional geometric models that do not erase reflections, and stereochemistry aware string representations. On the generative side, we look at how to penalize mirror duplicates, enforce stereochemical plausibility and embed chiral objectives. Throughout, ideas from Moores and Zuin Zeidler about epistemic responsibility serve as a background filter.
4.1 Data that tell the truth about stereochemistry
Everything downstream depends on how data are recorded and cleaned.
If a dataset does not distinguish racemates from enantiomers, and if stereochemistry is missing or contradicts the assay metadata, the best model in the world will learn a distorted picture.
For drug discovery work, a reasonable minimum standard looks like this.
- Racemates and enantiomers are registered as separate entities. A racemate gets its own identifier and carries metadata that it is a 50:50 mixture unless specified otherwise. Each enantiomer has its own record with explicit configuration.
- Assay records include which form was tested. If a racemate was used, the result is tagged accordingly. If an enantiopure sample was tested, that is stated clearly.
- Key chiral drugs and series are curated more carefully. For compounds such as omeprazole, citalopram, warfarin, propranolol and many others, data managers deliberately create stereo-specific entries and link them correctly to literature.
Public efforts like ChiralDB are beginning to assemble curated sets of stereochemically annotated molecules and reactions that can serve both as training data and as benchmarks for chiral-aware models. Updated versions of ChEMBL and similar resources place more emphasis on correct stereochemistry than earlier releases, although gaps and inconsistencies remain.
The point is not that a large organization can magically fix all legacy data. The point is that teams should know which parts of their data landscape are stereochemically trustworthy and which are not, and design models and decisions accordingly.
4.2 Representations that treat chirality as a first class feature
Once the data carry useful stereo-information, the model has to see it.
4.2.1 Two and a half dimensional graphs
A pragmatic path for many teams is to extend two dimensional graph neural networks with explicit stereo features. The idea is sometimes called “two and a half dimensional” because it adds three dimensional information in symbolic form without requiring full coordinates.
The recipe:
- Use standard tools such as RDKit to detect stereocenters and E or Z double bonds.
- Add node features indicating whether an atom is a stereocenter and, if so, its configuration in some consistent coding (for example, R or S).
- Add edge features to represent E or Z configuration on double bonds.
- Use these as part of the input feature set for a graph neural network.
Models like CageNet and related chirality aware GNNs show that this approach can significantly improve performance on tasks where stereochemistry matters, without incurring the cost of conformer generation for all molecules.
In this view, “is this atom chiral” and “what is its configuration” are as fundamental as “what is this atom’s element”.
4.2.2 Three dimensional models that keep track of handedness
For systems that already rely on three-dimensional conformers, it makes sense to use geometric models. The key design choice is whether the model treats reflections as a symmetry.
- If the model is invariant to translations and rotations but not to reflections, then enantiomers will map to different representations, as desired.
- If the model is invariant to reflections as well, then enantiomers collapse into the same point.
Architectures can be designed to be equivariant to the Euclidean group without including reflections, or to use oriented features such as signed volumes and oriented local frames that distinguish left handed arrangements from right handed ones.
You still have to worry about conformer generation quality, but at least the model has the capacity to learn chirality sensitive patterns such as enantioselective binding.
4.2.3 String representations that do not break under stereo
On the string side, SELFIES and Group SELFIES provide robust encodings that guarantee syntactic validity and can represent stereochemistry. In a stereochemistry aware generation setting:
- The alphabet includes tokens that encode R and S configurations and E or Z double bonds.
- The grammar ensures that generated strings respect valence and basic stereochemical rules.
- Models are trained with loss functions and objectives that do not push them to abandon rare stereo tokens simply because they are sparsely represented.
Tom and coworkers used such representations to build generative models that handle stereochemistry more faithfully than SMILES based baselines when tasks are explicitly chiral.
In short, if stability and robustness matter, you should not treat stereo tokens as exotic punctuation. They have to be integrated into the core alphabet and grammar that the model learns.
4.3 Generative constraints that keep output chemically honest
Even with clean data and good representations, generative systems need guard rails.
4.3.1 Penalising mirror duplicates and messy stereocenters
After molecules are generated, a clean up stage helps.
- Canonicalise structures with stereochemistry preserved.
- Detect mirror image pairs and decide whether to treat them as one design idea for the purpose of novelty metrics and prioritisation.
- Automatically filter out molecules with undefined stereocenters at positions where chirality is chemically crucial, unless the project explicitly wants to explore racemic mixtures.
- Reject structures that violate basic stereochemical feasibility.
This reduces the load on human reviewers and stops trivial mirror copies from flooding the top of the list.
4.3.2 Building stereo-aware rewards
For reinforcement learning style generative models, or any model that uses a reward based optimization loop, it is possible to build chiral awareness directly into the objective.
Examples:
- Use property predictors that distinguish enantiomers where relevant and include their outputs in the reward.
- When optimization is based on docking, use scoring functions and pose filters that are sensitive to the correct orientation of substituents in a chiral pocket.
- For tasks involving optical activity or enantioselective catalysis, use quantum chemical or experimental labels that depend on chirality.
Tom et al. construct this type of task in their PNAS Nexus work and show that stereo aware models can take advantage of the extra information.
4.3.3 Keeping Moores and Zuin Zeidler’s warning in sight
Moores and Zuin Zeidler argue that generative AI can easily start to dictate what looks like normal chemistry. They point out that language models can produce plausible but wrong molecular depictions and reaction mechanisms that, if left unchallenged, can reshape educational materials and intuition.
In the context of chiral bias, the practical way to honour this warning is to build a workflow where:
- AI generated structures are clearly labeled as such in internal tools and teaching material.
- Chemists are encouraged to question whether suggested stereochemistry and conformations actually make sense in the context of known mechanism and synthesis routes.
- Visual appeal of molecules is never accepted as a substitute for chemical validity.
The tool should supply candidates and visualisations. The decision about what counts as chemically real remains with the human experts.
4.4 Human in the loop checks
No matter how sophisticated the models become, stereochemical literacy in the human loop matters. Concrete checkpoints in a sensible pipeline might include:
- After virtual screening, a review to see whether the top ranked hits include many mirror image pairs. If so, decide explicitly which enantiomer to follow and which to discard.
- After generative design, a sanity check on the stereochemistry of top scoring candidates. Are crucial stereocenters specified How many suggestions rely on unrealistic chiral control
- Before key program decisions, a check that ADMET and tox models used for risk assessment treat known chiral drugs in a way that aligns with literature, rather than averaging.
Zuin Zeidler has stressed in interviews that laboratory experience and understanding of how data are produced are essential if chemists are to use AI safely. Without that, there is a tendency to assume that if a model draws a molecule or assigns a number, it must be correct.
Embedding explicit chiral sanity checks pushes back against that tendency.
4.5 Looking ahead
The path forward now has some structure.
- Data that correctly distinguish racemates and enantiomers and link labels to stereochemistry.
- Representations and models that treat chirality as a first class feature rather than an optional detail.
- Generative workflows that do not spam mirror duplicates or ignore stereochemical feasibility.
- Human and organizational habits that include chiral awareness by default.
The final episode steps back and asks what this looks like in a mature organisation and in education. It introduces the idea of chiral literacy as a cultural property, not just a technical one, and returns to Moores and Zuin Zeidler’s argument that generative AI should not define the visual and conceptual center of chemistry for the next generation of scientists.
References
Moores, Audrey & Zuin Zeidler, Vânia G. (2025). “Don’t let generative AI shape how we see chemistry.” Nature Reviews Chemistry, 9(10), 649–650. doi: 10.1038/s41570-025-00757-9
Tom G, Yu E, Yoshikawa N, Jorner K, Aspuru-Guzik A. Stereochemistry-aware string-based molecular generation. PNAS Nexus. 2025 Oct 14;4(11):pgaf329. doi: 10.1093/pnasnexus/pgaf329.
Kruithof, P.; et al. “Practical aspects of stereochemistry in cheminformatics and molecular modeling.”
Journal of Cheminformatics 2021, 13, 1–26. doi: 10.1186/s13321-021-00519-8
Fourches, D.; Muratov, E.; Tropsha, A. “Trust but verify: on the importance of chemical structure curation in cheminformatics.” J. Chem. Inf. Model. 2010, 50, 1189–1204. DOI: 10.1021/ci100176x
Gaulton A, et al. The ChEMBL database in 2023: a drug discovery platform and chemical biology resource. Nucleic Acids Research. 2023 51, D1282 to D1290.
Anderson BM, et al. CageNet: learning chirality aware graph representations for molecular property prediction. ChemRxiv preprint, 2021.
Zuin Zeidler, V. (2025, November 10). Chemistry and AI: Good data are gold [Interview]. Leuphana University Lüneburg. Retrieved from https://www.leuphana.de/en/institutions/faculty/sustainability/news/single-view/2025/11/10/chemistry-and-ai-good-data-are-gold.html
Krenn M, Haese F, Nigam AK, Friederich P, Aspuru Guzik A. Self referencing embedded strings SELFIES. Machine Learning: Science and Technology. 2020 1, 045024.
Schuett KT, Kindermans PJ, Sauceda HE, et al. SchNet: a continuous filter convolutional neural network for modeling quantum interactions. Journal of Chemical Theory and Computation. 2018 14, 6632 to 6642.
Bronstein MM, Bruna J, LeCun Y, Szlam A, Vandergheynst P. Geometric deep learning: grids, groups, graphs, geodesics and gauges. Nature Reviews Machine Intelligence. 2021 2, 743 to 755.
Walters WP. Applications of deep learning in molecule generation and molecular property prediction. Accounts of Chemical Research. 2021 54, 263 to 270.
Pollice R, et al. Data driven strategies for accelerated materials and molecular discovery. Accounts of Chemical Research. 2021 54, 849 to 860.
Yaëlle Fischer, Thibaud Southiratn, Dhoha Triki, Ruel Cedeno. Deep Learning vs Classical Methods in Potency & ADME Prediction: Insights from a Computational Blind Challenge. J. Chem. Inf. Model. 2025. https://doi.org/10.1021/acs.jcim.5c01982.
Schneider N, Lewis RA, Fechner N, Ertl P. Chiral Cliffs: Investigating the Influence of Chirality on Binding Affinity. ChemMedChem. 2018 Jul 6;13(13):1315-1324. doi: 10.1002/cmdc.201700798.
Husby J, Bottegoni G, Kufareva I, Abagyan R, Cavalli A. Structure-based predictions of activity cliffs. J Chem Inf Model. 2015 May 26;55(5):1062-76. doi: 10.1021/ci500742b.
Further Reading